Code organization
Nipoppy docs are moving
Nipoppy is undergoing a major refactor to move from scripts to a command-line interface (CLI) and Python API. The new documentation website (work in progress) can be found at https://nipoppy.readthedocs.io/.
If you are using the (soon-to-be legacy) scripts from Nipoppy 0.1.0, this is still the correct place to be. But we encourage you to check out the new website!
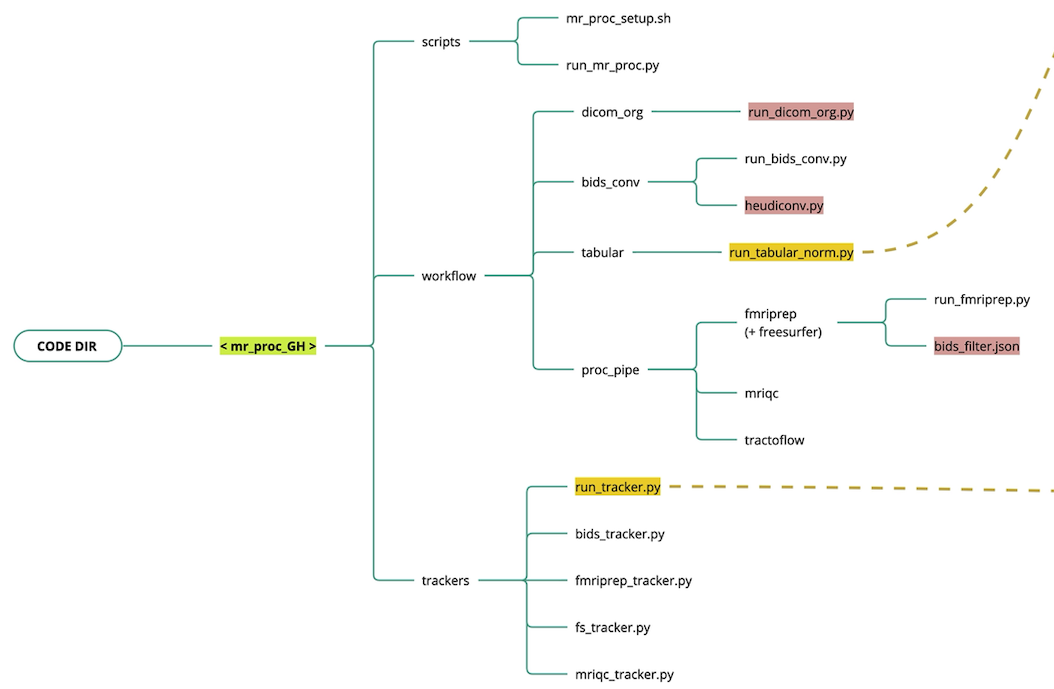
Code organization
The Nipoppy codebase is divided into data processing workflows and data availability trackers.
workflow
- MRI data organization (
dicom_organdbids_conv)- Custom script to organize raw DICOMs (i.e. scanner output) into a flat participant-level directory.
- Convert DICOMs into BIDS using Heudiconv
- MRI data processing (
proc_pipe)- Runs a set of containerized MRI image processing pipelines
- Tabular data (
tabular)- Custom scripts to organize raw tabular data (e.g. clinial assessments)
- Custom scripts to normalize and standardize data and metadata for downstream harmonization (see NeuroBagel)
trackers
- Track available raw, standardized, and processed data
- Generate
bagelsfor Neurobagel graph and dashboard.
Legend - Red: dataset-specific code and configuration files - Yellow: Neurobagel interface